OPTIMIZATION OF CRISPR/CAS12A TRANS-CLEAVAGE REACTION FOR DETECTION OF THE STAPHYLOCOCCUS AUREUS NUCA GENE
Institute of Tropical Medicine, Joint Vietnam-Russia Tropical Science and Technology Research Center
63 Nguyen Van Huyen, Nghia Do, Cau Giay, Ha Noi
Số điện thoại: +84974282734; Email: doai.vn@gmail.com
Nội dung chính của bài viết
Tóm tắt (Abstract)
Staphylococcus aureus (S. aureus) is the most prevalent pathogen within the staphylococcal group, classified as a gram-positive bacterium responsible for severe infections affecting the skin, bloodstream, organs, and gastrointestinal tract. Consequently, the development of early and accurate detection tools is a research priority. Recently, CRISPR/Cas technology, particularly Cas12a, has emerged as a promising diagnostic tool due to its ability to trans-cleave single-stranded DNA (ssDNA) following the specific cleavage of double-stranded DNA (dsDNA) targets. In this study, we optimized a CRISPR/Cas12a trans-cleavage reaction for detecting the nucA gene, a specific marker of S. aureus. To enhance enzyme activity, four crRNAs (crRNA1–4) were designed and tested, each exhibiting different effects on the reaction. However, a mixture of three crRNAs demonstrated higher efficiency than reactions using a single crRNA. Additionally, key reaction parameters, including enzyme concentration, additives, probe sequence, and incubation temperature, were systematically evaluated. The optimized reaction conditions included 6.25 nM LbCas12a combined with the mixture of three crRNAs (crRNA1–3) in a reaction containing 10 mM dithiothreitol (DTT) and 1 µM TA5C ssDNA FAM-BHQ1 probe. The assay was incubated at 39°C for 30 minutes, achieving a detection limit of 284.8 amol or 1.72 × 10⁸ copies of the nucA gene. Fluorescent signals were detectable under a standard UV transilluminator. This study provides valuable insights for the development of CRISPR-based diagnostic kits for S. aureus detection.
Từ khóa (Keywords)
Staphylococcus aureus, nucA, CRISPR/Cas12a, trans-cleavage, diagnostic
Chi tiết bài viết
Highlights
A combination of three crRNAs enhances the efficiency of CRISPR/Cas12a-based detection of the nucA gene, specific to Staphylococcus aureus.
Optimized detection parameters include 1.56–12.5 nM LbCas12a, a crRNA1–3 mixture, 10 mM dithiothreitol (DTT), and 1 µM TA5C ssDNA FAM-BHQ1 probe.
The optimized assay achieves a detection limit of 284.8 amol or 1.72 × 10⁸ copies of the nucA gene in a 20 µl reaction.
1. INTRODUCTION
Staphylococcus aureus (S. aureus) is a Gram-positive, facultative anaerobic bacterium and a major human pathogen within the Staphylococcus genus. It is commonly found as a commensal organism on the skin and mucous membranes but can become opportunistic, causing a wide spectrum of infections. These range from mild skin conditions, such as impetigo and abscesses, to severe and life-threatening diseases, including pneumonia, endocarditis, and sepsis [1].
A key factor in the pathogenicity of S. aureus is its ability to produce a variety of virulence factors, including toxins (e.g., enterotoxins, toxic shock syndrome toxin) and enzymes (e.g., coagulase, hyaluronidase), which facilitate tissue invasion and immune evasion [2]. S. aureus infections are particularly concerning in healthcare settings, where they are a leading cause of hospital-acquired infections, especially among immunocompromised patients [3]. Therefore, rapid and accurate detection methods are crucial for effective treatment and infection control.
In addition to traditional culture, biochemical, and immunological methods, molecular diagnostic methods have been developed for the detection of S. aureus [4]. These methods offer the advantage of rapid bacterial DNA detection in clinical, food, and environmental samples. They are often highly sensitive and specific, making them valuable tools in diagnostic applications. As a result, despite the widespread use of automated isolation, culture, and identification techniques, molecular detection kits for S. aureus continue to be developed and successfully commercialized.
Molecular detection methods, such as conventional PCR and real-time PCR, are typically designed to target genes specific to S. aureus, including the 16S rRNA gene, nuclease A (nucA), coagulase (coa), and protein A (spa) [5,6]. Among these, the nucA gene, which encodes a heat-stable nuclease capable of degrading nucleic acids, is widely used due to its specificity for S. aureus and its role in distinguishing certain Staphylococcus species [7,8].
CRISPR/Cas technology has recently been extensively researched and developed, demonstrating significant potential in gene editing and molecular diagnostics. This technology is based on the adaptive immune system of bacteria, with its core component being the CRISPR ribonucleoprotein (RNP) complex. The CRISPR RNP complex consists of Cas and guide RNAs, which enable the precise cleavage of target nucleic acids [9]. Acting as molecular scissors, Cas enzymes provide a highly specific, efficient, and versatile platform for genetic modification and diagnostic applications [10].
Similar to other systems, CRISPR/Cas12a is being developed for molecular diagnostic applications. Cas12a is guided by a single CRISPR RNA (crRNA) that recognizes a T-rich protospacer adjacent motif (PAM) sequence to cleave double-stranded DNA (dsDNA) targets. Specifically, LbCas12a requires a TTTV PAM sequence [11]. Notably, Cas12a exhibits trans-cleavage activity on single-stranded DNA (ssDNA) after being activated by a cis-cleavage reaction [12]. Under the guidance of crRNA, Cas12a binds to the target DNA, forming a ternary complex with crRNA and the target DNA. Once activated, Cas12a non-specifically cleaves ssDNA [12,13]. This property forms the basis of fluorescence-based detection: single-stranded DNA probes labeled with a fluorophore and quencher are added to the reaction. When cleaved by activated Cas12a, the fluorophore is separated from the quencher, producing a detectable fluorescence signal that can be visualized using a simple UV emitter [12].
CRISPR/Cas12a technology has been successfully applied to detect S. aureus [14–17]. These studies have developed methods that integrate CRISPR/Cas12a with isothermal amplification techniques such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) to target specific genes. However, the Cas12a reaction remains an active area of research, with ongoing optimizations. In this study, novel crRNAs were designed to specifically target the nucA gene of S. aureus and were combined with optimization strategies to enhance Cas12a trans-cleavage activity. The findings will contribute to the development of faster and more effective S. aureus diagnostic kits.
2. MATERIALS AND METHODS
2.1. Material
The S. aureus ATCC® 29213™ was purchased from American Type Culture Collection (ATCC.org).
2.2. CrRNA design
The nucA sequences of S. aureus and its orthologs were retrieved from GenBank (NCBI) and aligned to identify a species-specific region of the S. aureus nucA gene. Next, crRNA target sites were selected based on the presence of a specific PAM sequence (TTTV). Potential off-target were assessed using Cas-OFFinder [18] and BLAST-N (NCBI). The selected crRNAs were synthesized using the chemical RNA synthesis method (IDT).
2.3. Preparation of Target dsDNA
Staphylococcus aureus cells were suspended in 50 µL of 1X TE buffer and heated at 95ºC for 5 minutes. Then, 2 µL of the heat-treated solution was used for polymerase chain reaction (PCR). The PCR reaction mixture consisted of 25 µL of Taq 2X Ready Mix (Nippon Genetics), 1 µL each of 10 µM forward and reverse primers (nucA-F, nucA-R, Table 1), 2 µL of the heat-treated cell solution, and 22 µL of water.
Table 1. Oligonucleotide sequences used in this study
Oligo name | Sequence (5’ à 3’) | Oligo type | Reference |
rpa-nucF | AGCGATTGATGGTGATACGGTTAAATTAATGTAC | ssDNA primer | This study |
rpa-nucR | ACTTTAGCCAAGCCTTGACGAACTAAAGCTTCG | ssDNA primer | This study |
6T | FAM/TTTTTT/BHQ1 | ssDNA probe | [19] |
TA5C | FAM-TTATTCCCCC-BHQ1 | ssDNA probe | [20] |
crRNA1 | UAAUUUCUACUAAGUGUAGAUACAAAGGUCAAAGAACUGAUAAAUA | single-stranded RNA | This study |
crRNA2 | UAAUUUCUACUAAGUGUAGAUUGCUGAUGGAAAAAUGGUAAACGAA | single-stranded RNA | This study |
crRNA3 | UAAUUUCUACUAAGUGUAGAUAGGUGUAUCAACUAAUAAUAGUCUGA | single-stranded RNA | This study |
crRNA4 | UAAUUUCUACUAAGUGUAGAUACCUUUGUCAAACUCGACUUCAAUU | single-stranded RNA | This study |
The PCR cycling condition was as follows: initial denaturation at 95ºC for 3 minutes, followed by 35 cycles of denaturation at 95ºC for 20 seconds, annealing at 60ºC for 30 seconds, and a final extension at 72ºC for 5 minutes. The resulting amplicons were analyzed by agarose gel electrophoresis and purified using the GeneJET Gel Extraction Kit (Thermo Scientific). The purified DNA was quantified using a NanoDrop spectrophotometer and diluted to a final concentration of 1–500 pg.
2.4. Optimization of CRISPR/Cas12a trans-cleavage reaction
The CRISPR/Cas12a trans-cleavage assay was originally performed following an established protocol [21] with several modifications. Briefly, a 20 µL reaction was prepared in a 100 µL PCR tube containing 1X NEBuffer r2.1 (NEB), 0–50 nM LbCas12a (IDT), crRNA at twice the concentration of LbCas12a (crRNA 1–4, Table 1), 0–2 µM ssDNA probe (6T or TA5C, Table 1), 20 units of an RNA inhibitor (Meridian Bioscience), and 0–0.5 ng of target dsDNA.
The reaction was incubated at 32 - 41ºC in a CFX96 real-time PCR system (Bio-rad), and fluorescence signals (λexcitation: 493 nm; λemission: 517 nm) were automatically recorded every 30 seconds. All experiments were performed in triplicate, and data are presented as the mean ± standard deviation of relative fluorescence units (RFU).
For fluorescence imaging, tubes were placed on a UV transilluminator and photographed using a digital camera.
3. RESULTS
3.1. Designation and selection of crRNA
To design crRNAs, the nucA gene sequence was retrieved from the Staphylococcus aureus reference genome (GenBank: #NC_007795). Using BLASTN (NCBI), six orthologs of nucA were identified (Supplementary Figure F1). Based on sequence alignment, four crRNAs specific to nucA in S. aureus were selected (Table 1, Figure 1A, Supplementary Figure F1). No potential off-target effects were predicted by Cas-OFFinder or BLASTN (data not shown). Previous studies have shown that crRNA sequences can differentially influence Cas12a enzyme activity [19]. Therefore, these four crRNAs (crRNA1–4) were selected and chemically synthesized to identify potential candidate sequences.
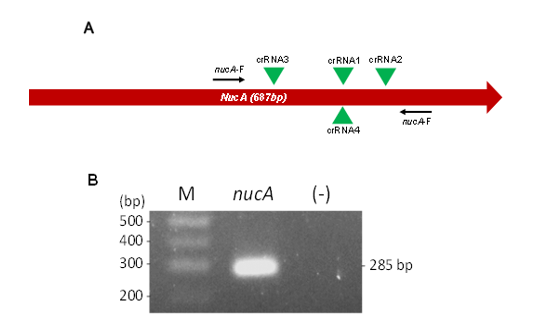
Figure 1. Design of crRNA and Preparation of Target dsDNA. (A) Schematic representation of the nucA gene, highlighting primer binding sites (arrows) and crRNA target regions (green triangles). (B) Agarose gel electrophoresis analysis of nucA amplicons. M. 100 bp DNA ladder; (-) negative control; bp: base pair
Next, the DNA regions encompassing the four crRNAs were amplified using PCR with specific primers (Table 1, Figure 1A) to generate dsDNA targets. Agarose gel electrophoresis confirmed successful and specific amplification, showing PCR products of approximately 285 bp (Figure 1B). The resulting purified amplicons were then used to evaluate crRNA performance in a Cas12a reaction. Initially, 0.1 ng of amplicons were added to each reaction. Under identical conditions, Cas12a with crRNA2 exhibited the highest trans-cleavage activity, as indicated by the most substantial fluorescence signal curve (Figure 2A). This was followed by crRNA1 and crRNA3, which showed intermediate activity levels, while crRNA4 had the lowest activity.
The combination of crRNAs has been shown to enhance trans-cleavage activity [22]. In this study, reactions using crRNA2 alone and a mixture of all three crRNAs (crRNA1–3) were evaluated. The crRNA4 was excluded due to its low activity and the proximity of its and crRNA1 target sequences. The results showed that the mixed crRNAs generated a higher fluorescence signal curve compared to the reaction utilizing crRNA2 alone (Figure 2B). Therefore, the reaction with mixed crRNAs (crRNA123) was used for subsequent experiments.
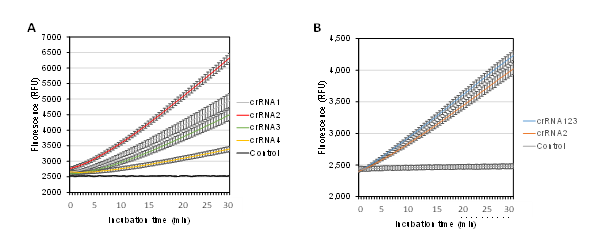
Figure 2. Effect of crRNAs on LbCas12a activity. Fluorescent signal from Cas12a trans-cleavage reactions using different single crRNAs (A) and a mixture of four crRNAs (B). Control: no cRNA added
3.2. Effect of Chemical Enhancers on Trans-Cleavage Activity
To evaluate the impact of chemical enhancers on trans-cleavage activity, 10 mM DTT was added to the reaction mixture, as previous studies have suggested its potential to enhance ssDNA probe cleavage [23]. Reactions were performed with and without DTT to assess its effect under the current buffer conditions. The results showed that the fluorescence signal was significantly higher in the DTT-containing reaction compared to the control (Figure 3). Based on this observation, DTT was included in subsequent optimization steps.
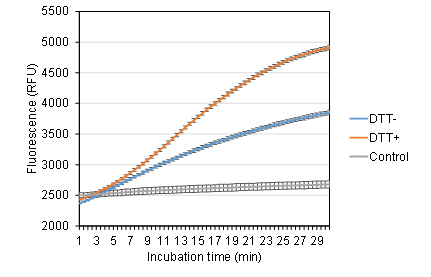
Figure 3. Effect of DTT on LbCas12a activity. Reactions were performed in NEBuffer r2.1 with or without 10 mM DTT (DTT+ and DTT−, respectively). Control: No DNA target
3.3. Optimization of LbCas12a Trans-Cleavage Reporters
The sequence of the ssDNA probe influences the trans-cleavage activity of Cas12a [19]. In this study, two probe sequences, TA5C and 6T, were evaluated by comparing fluorescence emission from Cas12a-mediated ssDNA trans-cleavage. The probes were designed as ssDNA oligonucleotides with FAM at the 5′ end and BHQ1 at the 3′ end. Cleavage of the ssDNA can release the FAM which generates fluorescence signal. The results showed that the TA5C probe generated a higher fluorescence signal at an earlier stage of the reaction, making it the preferred choice for further studies (Figure 4A).
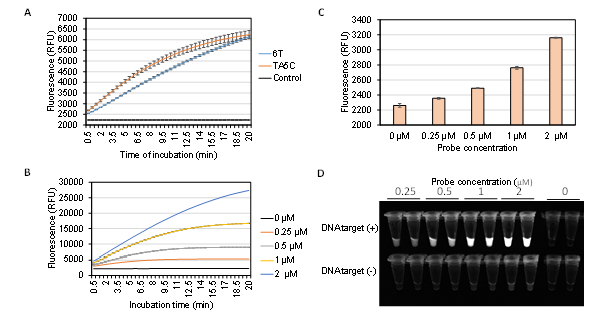
Figure 4. Influence of ssDNA probe on LbCas12a-specific trans-cleavage activity. (A) Effect of ssDNA probe sequence on trans-cleavage activity. Control: no ssDNA probe added. (B) Impact of probe concentration on trans-cleavage efficiency. (C) Effect of probe concentration on fluorescence background. (D) Fluorescence images of incubated tubes with (+) or without (-) target DNA
Additionally, the concentration of the ssDNA probe affects the signal intensity [19]. To optimize this parameter, experiments were conducted using five different probe concentrations (0–2 µM). The results indicated that fluorescence intensity increased proportionally with probe concentration (Figure 4B,D). However, higher probe concentrations also led to increased background fluorescence in control reactions lacking a dsDNA target (Figure 4C,D). Under UV light, the fluorescence signal in control reactions with a 2 µM probe was detectable by the naked eye, which could impact future diagnostic applications. Therefore, a concentration of 1 µM was selected for subsequent experiments.
3.4. Optimization of Incubation Temperature
Temperature generally influences the rate of enzymatic reactions. In this step, the effect of incubation temperature on the Cas12a-catalyzed trans-cleavage reaction was investigated. The earliest fluorescence signal curves were observed at 39 and 40°C. In contrast, reactions at lower temperatures (32–37°C) proceeded more slowly but reached higher maximum fluorescence intensities. Notably, temperatures above 40°C had a negative impact on the reaction, resulting in delayed signal curves and reduced maximum fluorescence intensity (Figure 5).
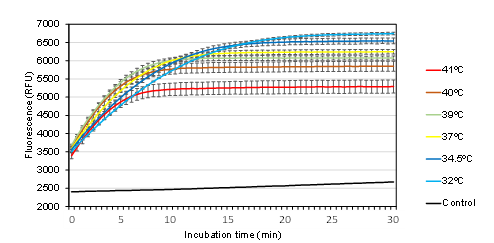
Figure 5. Correlation between relative fluorescence units (RFUs) and incubation temperature. Control: reaction at 39ºC without a dsDNA target
In this study, incubation temperature of 39°C was selected to align with isothermal amplification reactions and, in particular, to optimize reaction time.
3.5. Selection of Enzyme Concentration
Enzyme concentration is a key factor influencing reaction rates. To determine the optimal conditions, LbCas12a was tested at concentrations ranging from 0 to 50 nM. Additionally, the crRNA mixture was added at a twofold molar excess relative to LbCas12a. The fluorescence signal curve gradually improved with increasing enzyme concentrations from 0.1 to 12.5 nM (Figure 6). However, no significant difference was observed between reactions containing LbCas12a at 1.56–12.5 nM. In contrast, higher enzyme concentrations (25 and 50 nM) negatively affected the fluorescence signal. Based on these findings, the optimal concentration range for LbCas12a was determined to be between 1.56 and 12.5 nM. A mid-range concentration of 6.25 nM was selected as the working condition for subsequent experiments.
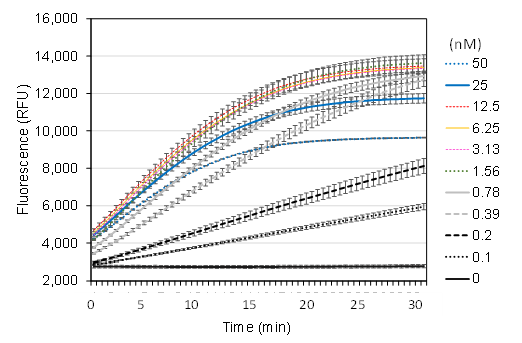
Figure 6. Impact of LbCas12a enzyme concentration on trans-cleavage activity
3.6. Detection Limit of the CRISPR-based detection system
To determine the detection limit of the nucA gene using CRISPR-based diagnostics, reactions were performed with varying amounts of dsDNA template (0–500 pg) under the optimized conditions. The enzyme concentration was set at 6.25 nM, the average value within the recommended range. Fluorescence signal curves were recorded for reactions containing 50–500 pg of dsDNA target (Figure 7A). Additionally, fluorescence signals after a 30-minute reaction were visually assessed under ultraviolet (UV) light (Figure 7B). In reactions with lower dsDNA target concentrations, no distinguishable fluorescence was observed compared to the control.
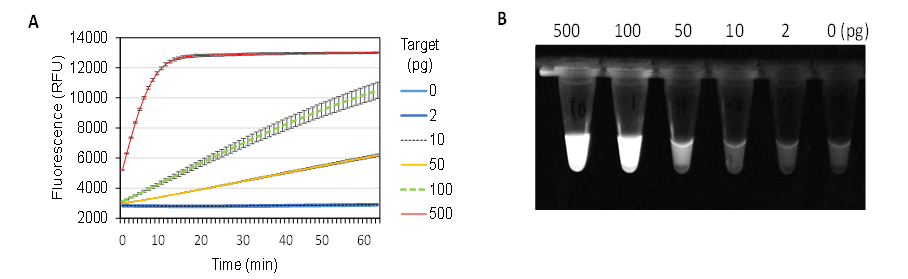
Figure 7. Detection Limit of the CRISPR/Cas12a Trans-Cleavage Reaction for the Staphylococcus aureus nucA Gene. (A) Real-time fluorescence signal. (B) Fluorescence image of tubes under UV light. The reactions were incubated at 39°C for 30 min before imaging
Thus, under optimized conditions, the CRISPR-based detection system demonstrated a sensitivity limit of at least 50 pg of the nucA gene segment (285 bp), corresponding to approximately 284.8 amol or 1.72 × 108 copies.
4. DISCUSSION
4.1. Effect of Reaction Components on the Trans-Cleavage Activity of LbCas12a
The composition of reaction components plays a crucial role in determining the rate and efficiency of the trans-cleavage reaction. Factors such as crRNA and probe sequences, enzyme and probe concentrations, buffer conditions, and chemical additives significantly influence overall reaction performance [19]. Optimizing these parameters is essential to maximize LbCas12a activity and improve assay sensitivity and specificity.
In this study, the results demonstrated that the enzyme complex with crRNA2 exhibited higher activity compared to other crRNAs (Figure 2A). Additionally, the reaction using a combination of crRNAs (crRNA1–3) showed greater trans-cleavage activity than reactions with a single crRNA (Figure 2B). This finding aligns with previous studies, suggesting that the use of multiple crRNAs increases the number of cleavage sites per DNA molecule, thereby enhancing LbCas12a activation and improving overall trans-cleavage efficiency [22].
The sequence and concentration of probes significantly influence the reaction rate and fluorescence signal intensity. Previous studies have shown that TA5C probes outperform several other sequences in cleavage efficiency [20]. In this study, reactions using TA5C probes consistently demonstrated higher cleavage efficiency than those using 6T probes (Figure 4A). Additionally, increasing the probe concentration enhanced fluorescence signal intensity; however, it also elevated background fluorescence. While background signals can be mitigated using negative controls and fluorescence spectrometers, they may still interfere with results when assessed visually. Notably, the Cas12a system has shown potential for operation in minimal laboratory settings, utilizing conventional UV transiluminator systems for detection.
The addition of DTT has been shown to enhance the trans-cleavage activity of Cas12a [23]. Consistently, in this study, DTT supplementation significantly increased the fluorescence signal, nearly doubling that of the control (Figure 3). These results highlight the beneficial effect of chemical additives in improving reaction efficiency. Further research is needed to identify additional enhancers that can further optimize the performance of the Cas12a system.
Finally, enzyme concentration is critical in optimizing the Cas12a diagnostic system. The results indicated that the optimal concentration range was 1.56–12.5 nM (Figure 6). However, this outcome is highly dependent on enzyme quality and storage conditions. Therefore, to ensure consistent performance, the protein concentration must be re-optimized for each new batch of enzymes. Notably, increasing the enzyme concentration above 25 nM had a negative effect on the reaction, though the underlying mechanism remains unclear.
4.2. Effect of Reaction Conditions on the Trans-Cleavage Activity of LbCas12a
Temperature influences the reaction rate of most enzymes, including Cas12a. While Cas12a remains active at lower temperatures, optimal activity requires temperatures of 28°C or higher [24]. Although Cas12a can tolerate temperatures up to 52°C [25], the results of this study showed that temperatures above 40°C reduced enzyme activity (Figure 5). Notably, while lower temperatures slowed the reaction rate, they resulted in higher maximum fluorescence intensity. This suggests that enzyme stability may be better maintained at lower temperatures, leading to more sustained activity over time. Therefore, the optimal temperature should be selected based on the specific application, considering factors such as compatibility with other reactions, time constraints, and the desired signal intensity.
4.3. Detection Limit of the optimized amplification free CRISPR-based detection system
Although Cas12a is regarded as an accurate and efficient detection tool, its sensitivity is typically limited to the picomolar range without amplification or signal enhancement strategies [26,27]. In this study, the optimized CRISPR diagnostic system achieved a detection limit of approximately 284.8 amol or 1.72 × 10⁸ copies of the nucA gene per 20 µL reaction, corresponding to a target concentration of 14.24 fM. This result aligns with previous findings, such as a system capable of detecting as little as 25 fM of PCA3 gene amplicons [27]. Consequently, integrating pre-amplification or signal amplification techniques is crucial for enhancing sensitivity and broadening the system's applicability in diagnostic settings. For instance, combining Recombinase Polymerase Amplification (RPA) with the CRISPR/Cas12a system has achieved a detection limit of 10 genome copies per reaction [21]. Recently, post-reaction amplification technologies have been developed with exceptional sensitivity. For example, amplification systems using cascade reactions can achieve detection limits as low as one to a few genome copies per μL of reaction [28,29]. These advanced amplification strategies can be fully integrated with the optimised Cas12a system from this study to develop comprehensive diagnostic kits.
5. CONCLUSION
In this study, key parameters were optimized for the CRISPR-based detection of the nucA gene, a specific marker of S. aureus. The optimized reaction included 6.25 nM LbCas12a combined with a mixture of three crRNAs (crRNA1–3) in a buffer containing 10 mM DTT and 1 µM TA5C ssDNA FAM-BHQ1 probe. Under incubation at 39°C for 30 minutes, the amplification-free CRISPR assay achieved a detection limit of 284.8 amol or 1.72 × 10⁸ copies of the nucA gene per 20 µL reaction. These findings offer valuable insights for the development of S. aureus detection kits, paving the way for more rapid and sensitive diagnostic methods.
Acknowledgement: This research was supported by project “Development of an RPA-CRISPR/Cas12-based detection method for S. aureus and MRSA”. Funding was provided by Joint Vietnam-Russia Tropical Science and Technology Research Center.
Author contributions: Nguyen Van Doai, Ta Thi Loan, Le Thi Lan Anh conceived and supervised the study. Nguyen Giang Son designed the crRNAs. Nguyen Van Doai and Nguyen Phuong Thuy performed the experiments. Nguyen Van Doai analyzed and presented the data. Nguyen Van Doai wrote the manuscript. Nguyen Van Doai, Hoang Dang Hieu, Le Thi Van Anh, Le Thi Lan Anh revised the manuscript. All authors read and approved the final manuscript.
Conflicts of interest statement: The authors report no financial or any other conflicts of interest in this work.
Supplementary information (Supplementary Figure F1)
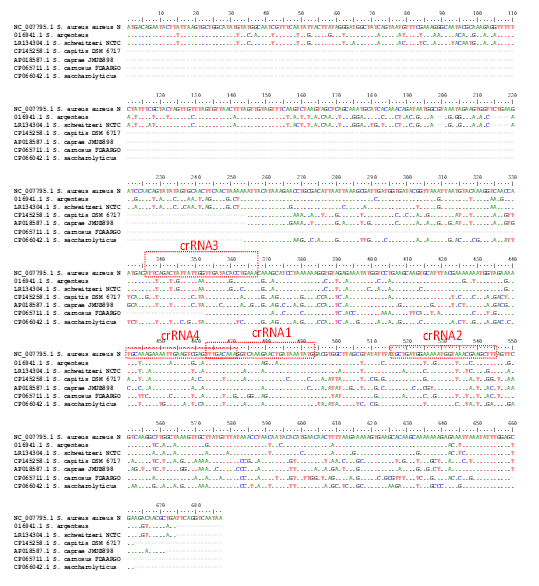
Supplementary Figure F1. Sequence alignment of nucA gene sequences. The boxed regions indicate crRNA target sites.
Tài liệu tham khảo
2. G.Y.C. Cheung, J.S. Bae and M. Otto, Pathogenicity and virulence of Staphylococcus aureus, Virulence, Vol.12, pp. 547–569, 2021. DOI: 10.1080/21505594.2021.1878688.
3. A.G. Jensen et al. , risk factors for hospital-acquired Staphylococcus aureus bacteremia, Arch. Intern. Med., Vol. 159, pp. 1437, 1999. DOI:10.1001/archinte.159.13.1437
4. A. Sanchini, Recent developments in phenotypic and molecular diagnostic methods for antimicrobial resistance detection in Staphylococcus aureus: A Narrative Review, Diagnostics, Vol.12, pp. 208, 2022. DOI: 10.3390/diagnostics12010208
5. F.-J. Schmitz, et al., Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by a multiplex PCR, Journal of Medical Microbiology, Vol.46, pp. 773–778, 1997. DOI: 10.1099/00222615-46-9-773
6. W.J. Mason, J.S. Blevins, K. Beenken, N. Wibowo, N. Ojha and M.S. Smeltzer, Multiplex PCR protocol for the diagnosis of Staphylococcal infection, J. Clin. Microbiol., Vol.39, pp. 3332–3338, 2001. DOI: 10.1128/JCM.39.9.3332-3338
7. J. Tang et al., The staphylococcal nuclease prevents biofilm formation in Staphylococcus aureus and other biofilm-forming bacteria, Sci. China Life Sci., Vol.54, pp. 863–869, 2011. DOI: 10.1007/s11427-011-4195-5
8. K. Tam, V.J. Torres, Staphylococcus aureus secreted toxins and extracellular enzymes, Microbiol. Spectr., Vol.7, pp. 7.2.16, 2019. DOI: 10.1128/microbiolspec.GPP3-0039-2018
9. R. Barrangou, The roles of CRISPR–Cas systems in adaptive immunity and beyond, Current Opinion in Immunology, 2015. Vol.32, pp. 36–41. DOI: 10.1016/j.coi.2014.12.008
10. G.A. Heinz, M.-F. Mashreghi, CRISPR-Cas-System als molekulare schere für gentherapie, Z. Rheumatol., Vol.76, pp. 46–49, 2017. DOI: 10.1007/s00393-017-0267-7
11. E. Tóth et al., Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases, Nucleic Acids Research, Vol.48 ,pp. 3722–3733, 2020. DOI: 10.1093/nar/gkaa110
12. S.-Y. Li, Q.-X. Cheng, J.-K. Liu, X.-Q. Nie, G.-P. Zhao and J. Wang, CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA, Cell Res, Vol.28, pp. 491–493, 2018. DOI: 10.1038/s41422-018-0022-x
13. J.S. Chen et al., CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity, Science, Vol.360, pp.436–439, 2018. DOI: 10.1126/science.aar6245
14. X. Cao et al., Cas12a/guide RNA-based platforms for rapidly and accurately identifying Staphylococcus aureus and methicillin-resistant S. aureus, Microbiol. Spectr. , Vol.11, pp. e04870-22, 2023. DOI: 10.1128/spectrum.04870-22
15. D. Xu, H. Zeng, W. Wu, H. Liu and J. Wang, Isothermal amplification and CRISPR/Cas12a-system-based assay for rapid, sensitive and visual detection of Staphylococcus aureus, Foods, Vol.12, pp. 4432, 2023. DOI: 10.3390/foods12244432
16. M. Zeinulin, M. Amanzholova, A. Shaizadinova and S. Abeldenov, Advancement in Staphylococcus aureus detection using a RPA-CRISPR-Cas12a fluorescent assay technology, Eurasian Journal of Applied Biotechnology,pp. 23–28, 2023. DOI: 10.11134/btp.3.2023.3
17. Y. Liu et al., One-tube RPA-CRISPR Cas12a/Cas13a rapid detection of methicillin-resistant Staphylococcus aureus, Analytica. Chimica. Acta., Vol.1278 , pp. 341757, 2023.DOI: 10.1016/j.aca.2023.341757
18. S. Bae, J. Park and J.-S. Kim, Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases, Bioinformatics, Vol.30, pp. 1473–1475, 2014. DOI: 10.1093/bioinformatics/btu048
19. H. Lv et al., Definition of CRISPR Cas12a trans-cleavage units to facilitate CRISPR diagnostics, Front. Microbiol, Vol.12, pp. 766464, 2021. DOI: 10.3389/fmicb.2021.766464
20. S. Lee et al., Highly efficient DNA reporter for CRISPR/Cas12a-based specific and sensitive biosensor, BioChip J., Vol.16, pp. 463–470, 2022. DOI: 10.1007/s13206-022-00081-0
21. Y. Li et al., Rapid one-tube RPA-CRISPR/Cas12 detection platform for methicillin-resistant Staphylococcus aureus, Diagnostics, Vol.12 , pp. 829, 2022. DOI: 10.3390/diagnostics12040829
22. Y. Chen et al., Multiple crRNAs-assisted CRISPR/Cas12a assay targeting cytochrome b gene for amplification-free detection of meat adulteration, Analytica Chimica Acta, Vol.1231, pp. 340417, 2022. DOI: 10.1016/j.aca.2022.340417
23. F. Deng, Y. Li, B. Li and E.M. Goldys, Increasing trans-cleavage catalytic efficiency of Cas12a and Cas13a with chemical enhancers: Application to amplified nucleic acid detection, Sensors and Actuators B: Chemical, Vol.373, pp. 132767, 2022. DOI: 10.1016/j.snb.2022.132767.
24. A.A. Malzahn et al., Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis, BMC Biol., Vol.17, pp. 9, 2019. DOI: 10.1186/s12915-019-0629-5
25. R.T. Fuchs et al., Characterization of Cme and Yme thermostable Cas12a orthologs, Commun. Biol., Vol.5, pp. 325, 2022. DOI: 10.1038/s42003-022-03275-2
26. H. Li et al., Amplification-free CRISPR/Cas detection technology: challenges, strategies, and perspectives, Chem. Soc. Rev., Vol.52, pp. 361–382, 2023. DOI: 10.1039/D2CS00594H
27. L.T. Nguyen, B.M. Smith and P.K. Jain, Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection, Nat. Commun., Vol.11, pp. 4906, 2020. DOI: 10.1038/s41467-020-18615-1
28. K. Shi et al., A CRISPR-Cas autocatalysis-driven feedback amplification network for supersensitive DNA diagnostics, Sci. Adv., Vol.7, pp.eabc7802, 2011. DOI: 10.1126/sciadv.abc7802
29. H. Sun et al., A programmable sensitive platform for pathogen detection based on CRISPR/Cas12a -hybridization chain reaction-poly T-Cu, Analytica Chimica Acta, Vol.1317, pp.342888, 2024. DOI: 10.1016/j.aca.2024.342888